COMBINE 2023

NEWS October 4:
The “Computational Modeling in Biology” Network (COMBINE) is an initiative to coordinate the development of the various community standards and formats in systems biology and related fields. COMBINE 2023 was a workshop-style event hosted at the Center for Cell Analysis and Modeling at the University of Connecticut School of Medicine, Farmington, CT, USA. The meeting was held in October 2023, closely aligned with the dates of the International Conference on Systems Biology (ICSB) October 8-12 in Hartford, CT. Tutorials at the last day (Sunday October 8th) took place at the ICSB 2023 venue - in the convention center. The meeting days included talks about the COMBINE standards and associated or related standardization efforts, presentations of tools using these standards, breakout sessions for detailed discussions as well as tutorials. There were no dedicated poster sessions, but participants were encouraged to bring posters - poster boards were provided next to meeting places. Some time each day was left for community discussion and wrap-ups of breakouts and advertisements for following breakouts. It was primarily an in-person meeting, with individual breakout sessions responsible for enabling remote participation as needed.
Local organizers were Michael Blinov (blinov@uchc.edu) and Ion Moraru (moraru@uchc.edu).
A tentative schedule
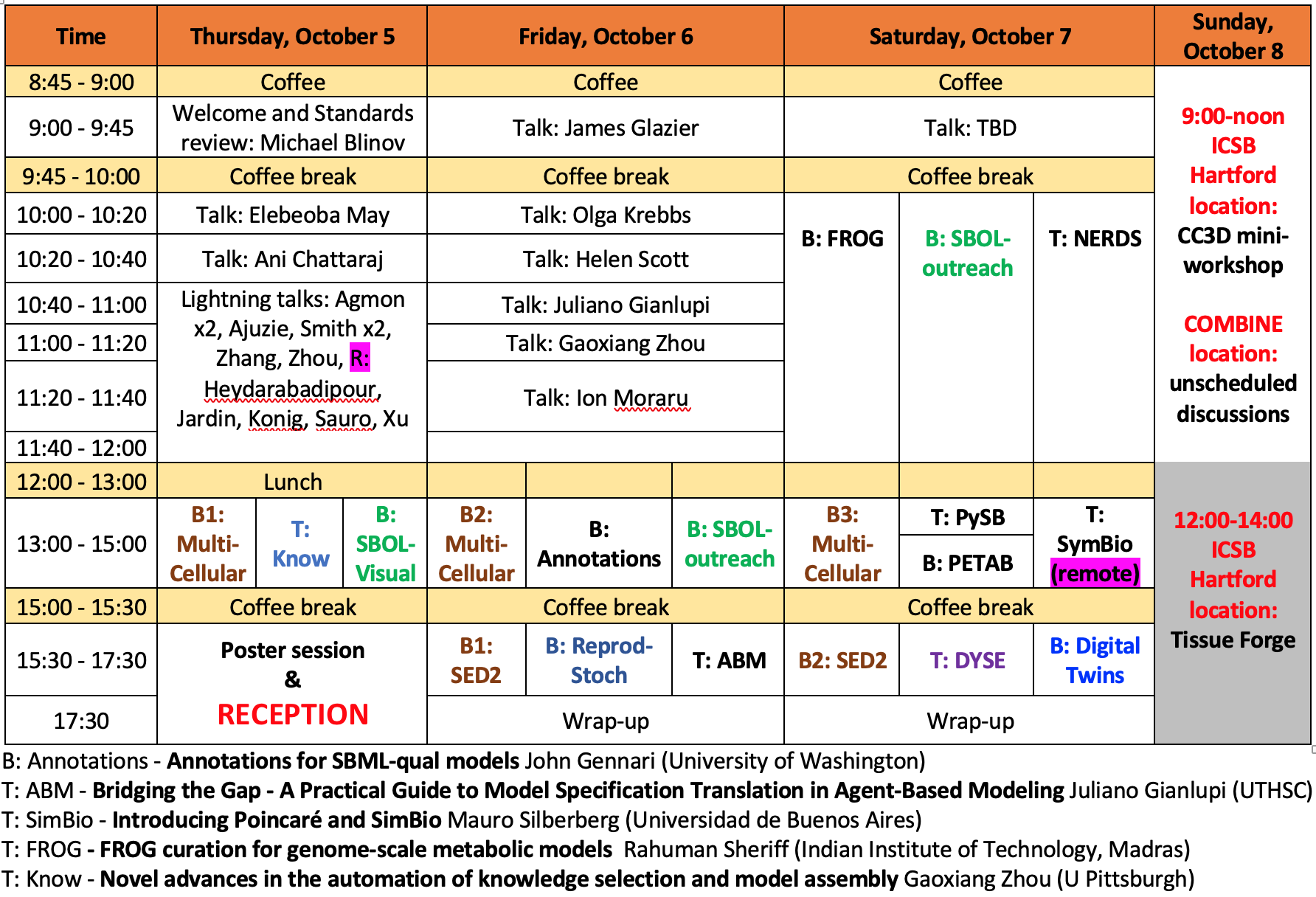 Note that many events are scheduled somewhat spontaneously at these events; keep an eye out here or on the COMBINE slack for last-minute changes and additions.
Note that many events are scheduled somewhat spontaneously at these events; keep an eye out here or on the COMBINE slack for last-minute changes and additions.
Workshop Location
COMBINE 2023 took place at the Center for Cell Analysis and Modeling (CCAM) in Farmington, CT. COMBINE 2023 took place in the Cell and Genome Sciences building, at 400 Farmington Ave, Farmington, CT 06119.

Arrival and Transportation
The closest airport is the Bradley International Airport (BDL). The recommended transportation is to take Uber/Lyft to 400 Farmington Ave, Farmington, CT. It takes about 25 minutes and costs about $40. You may rent a car at BDL - all parking at the workshop location and hotel are free. A less expensive but long travel from BDL is to take Bradley Flyer Bus (#30) from BDL to Central Row North Side at Old State House Station, and then take a bus 66T from Main St & Asylum St to 400 Farmington Ave. It takes 1 to 3 hours depending on schedules.
Accomodations and meals
The closest hotel to the CCAM is the Homewood Suites by Hilton Hartford-Farmington at 2 Farm Glen Boulevard, Farmington, Connecticut, 06032; it’s 7 minutes walk between there and the venue. A University rate of $154/night will be provided upon request. There are more hotels in the area, but any other hotel will require a car.
The hotel serves hot breakfast. There is a Butchers and Bakers restaurant 15 minutes walk, but no food stores within walking distance. We will serve food at the venue, depending on the sponsors it may be free or for a nominal fee.
Attendees
76 registered participants as of October 2
| Name Affiliation | Attendance in person | Interested Projects |
|---|---|---|
| Alan Garny University of Auckland | remotely | SED-ML, OMEX, COMBINE;CellML;Ontologies (SBO, KiSAO) libOpenCOR (https://opencor.ws/libopencor/) and OpenCOR (https://opencor.ws/) |
| Alex Patrie UConn Health | Oct 5 Oct 6 Oct 7 Oct 8 | SBML;SED-ML, OMEX, COMBINE;CellML;Multicellular modeling - |
| Amin Boroomand Woods Hole institute | Oct 5 Oct 6 Oct 7 Oct 8 | SBML;Multicellular modeling - |
| Amir Mahari University of Arkansas | Oct 5 Oct 6 Oct 7 Oct 8 | SBML;SBGN;Multicellular modeling I focus on the modeling and simulation of intercellular signaling pathways within melanoma cancer cells. This study holds significance due to the intricate nature of signaling cascades that regulate various cellular processes in melanoma progression. To facilitate this investigation, I employ PySB (Python Systems Biology), a powerful computational framework designed for creating, simulating, and analyzing mathematical models of biochemical systems. PySB’s modular and user-friendly nature enables me to construct complex models of intercellular signaling, incorporating factors such as ligand-receptor interactions, protein phosphorylation, and gene expression. Through PySB’s simulation capabilities, I can dynamically analyze the behavior of these pathways under different conditions, gaining insights into the underlying mechanisms that drive melanoma growth and metastasis. Ultimately, this research using PySB contributes to an enhanced understanding of melanoma biology, potentially paving the way for novel therapeutic interventions and personalized treatment strategies in combating this aggressive form of cancer. |
| Aniruddha Chattaraj Harvard University | Oct 5 Oct 6 Oct 7 Oct 8 | SED-ML, OMEX, COMBINE;Multicellular modeling Modeling of multivalent protein clustering, statistical analysis of cluster properties and visualization. Developer of MolClustPy. Interested in multi-scale modeling, specially problems related to biofilm formation. Software - Bionetgen, Virtual Cell, SpringSaLaD, LAMMPS, Python |
| Augustin Luna Harvard Medical School | remotely | SBGN;BioPAX https://www.pathwaycommons.org/; https://biofactoid.org/ |
| Bartholomew Jardine University of Washington | remotely | SBML;SED-ML, OMEX, COMBINE;Ontologies (SBO, KiSAO);Multicellular modeling Software tools for creating, editing, and simulating SBML complient modelss |
| Carolus Vitalis University of Colorado Boulder | SBOL, SBOL visual;Multicellular modeling - | |
| Chris Myers University of Colorado Boulder | remotely | SBML;SED-ML, OMEX, COMBINE;SBOL, SBOL visual SynBioHub/SynBioSuite/iBioSim |
| Dagmar Waltemath University Medicine Greifswald | remotely | SED-ML, OMEX, COMBINE;Ontologies (SBO, KiSAO) - |
| Dan Vasilescu UCHC | Oct 5 Oct 6 Oct 7 Oct 8 | SBML;SED-ML, OMEX, COMBINE;BioPAX;Ontologies (SBO, KiSAO) - |
| Daniel Ajuzie undergraduate | Oct 5 Oct 6 Oct 7 Oct 8 | SBML;SED-ML, OMEX, COMBINE;SBOL, SBOL visual;CellML;Ontologies (SBO, KiSAO);Multicellular modeling Ensemble Modeling; Model Optimization and Calibration |
| David Nickerson Auckland Bioengineering Institute, University of Auckland | remotely | SED-ML, OMEX, COMBINE;CellML;Ontologies (SBO, KiSAO);OMEX Metadata; FAIR indicators for models; repositories - |
| Diego Jahn TUD Dresden University of Technology, Center for Information Services and High Performance Computing (ZIH) | remotely | Multicellular modeling MorpheusML, MorpheusML Model Repository (https://morpheus.gitlab.io), Multicellular Modeling |
| Difei Tang University of Pittsburgh | Oct 5 Oct 6 Oct 7 Oct 8 | SBML;BioPAX;SBOL, SBOL visual;Ontologies (SBO, KiSAO) GUI for DySE framework |
| Dilan Pathirana University of Bonn | Oct 5 Oct 6 Oct 7 Oct 8 | SBML;Ontologies (SBO, KiSAO);Multicellular modeling;PEtab PEtab ( https://github.com/PEtab-dev/petab ). PEtab for model selection ( https://github.com/PEtab-dev/petab_select ). Model collection for benchmarking studies ( https://github.com/Benchmarking-Initiative/Benchmark-Models-PEtab/ ). |
| Edwin Moses Appiah Center for Cell Analysis and Modelling | Oct 5 Oct 6 Oct 7 Oct 8 | CellML;Multicellular modeling - |
| Egils Stalidzans PhD | Oct 5 Oct 6 Oct 7 Oct 8 | SBML;SBOL, SBOL visual ODE-based kinetic modeling, constraint based stoichiometric modeling |
| Elebeoba May University of Wisconsin-Madison | Oct 5 Oct 6 Oct 7 Oct 8 | SBML;Multicellular modeling;Visualization biochemical/bionetwork ODE models (metabolism, signal transduction, gene networks), multiscale models of host-pathogen systems, multiscale models of microbial communities, modeling synthetic bio systems |
| Eran Agmon UConn Health | Oct 5 Oct 6 Oct 7 Oct 8 | SED-ML, OMEX, COMBINE;Multicellular modeling Vivarium: https://vivarium-collective.github.io |
| Fengkai Zhang NIH | Oct 5 Oct 6 Oct 7 Oct 8 | SBML;SED-ML, OMEX, COMBINE;SBGN Simmune (https://www.niaid.nih.gov/research/simmune-project), rule-based modeling and libSBML-multi |
| Frank T. Bergmann BioQUANT, Heidelberg University | Oct 5 Oct 6 Oct 7 Oct 8 | SBML;SED-ML, OMEX, COMBINE;SBGN COPASI (https://copasi.org), basico (https://basico.readthedocs.io/), libSBML / libSEDML / libCombine |
| Gaoxiang Zhou University of Pittsburgh | Oct 5 Oct 6 Oct 7 Oct 8 | SBML;BioPAX;SBOL, SBOL visual;CellML;Multicellular modeling https://www.nmzlab.pitt.edu/people/gaoxiang-zhou, https://github.com/pitt-miskov-zivanov-lab, https://melody-biorecipe.readthedocs.io |
| Gerhard Mayer HITS (Heidelberg Institute for Theoretical Studies) gGmbH, Heidelberg | remotely | SED-ML, OMEX, COMBINE;Multicellular modeling EDITH (Ecosystem Digital Twins in Healthcare); https://www.edith-csa.eu |
| Herbert M Sauro University of Washington | remotely | SBML;SED-ML, OMEX, COMBINE;Multicellular modeling SBML, roadrunner etc |
| Ion Moraru UConn Health | Oct 5 Oct 6 Oct 7 Oct 8 | SBML;SED-ML, OMEX, COMBINE;model credibility VCell, BioSimulations, BioSimulators |
| Jacob Beal Raytheon BBN | Oct 5 Oct 6 Oct 7 Oct 8 | SBOL, SBOL visual - |
| James A. Glazier Indiana University | Oct 5 Oct 6 Oct 7 Oct 8 | Multicellular modeling CompuCell3D (https://compucell3d.org/), Virtual Cornea, IMAG/MSM Working Group on Multiscale Modeling and Viral Pandemics (https://www.imagwiki.nibib.nih.gov/working-groups/multiscale-modeling-and-viral-pandemics) |
| Jessica Yu Allen Institute for Cell Science | remotely | SED-ML, OMEX, COMBINE;Multicellular modeling agent-based modeling, cloud-based simulation and analysis workflows |
| Jim Schaff Contractor - UConn Health | Oct 5 Oct 6 Oct 7 Oct 8 | SED-ML, OMEX, COMBINE;Multicellular modeling Virtual Cell Project (vcell.org), Reproducible Biological Modeling (reproduciblebiomodels.org) |
| John Gennari University of Washington | Oct 6 Oct 7 | SBML;SED-ML, OMEX, COMBINE;BioPAX;CellML;Ontologies (SBO, KiSAO) Center for Reproducible Biomedical Modeling |
| Juliano Ferrari Gianlupi Postdoctoral Scholar, UTHSC | Oct 5 Oct 6 Oct 7 | SBML;CellML;Multicellular modeling PhenoCellPy https://www.biorxiv.org/content/10.1101/2023.04.12.535625v2.abstract; Translating PhysiCell specification into CompuCell3D simulation https://github.com/JulianoGianlupi/pcxml2cc3d |
| Jörn Starruß Technische Universität Dresden, Germany | remotely | SBML;SED-ML, OMEX, COMBINE;CellML;Multicellular modeling MorpheusML, https://morpheus.gitlab.io, Multicellular modeling, https://MultiCellML.org, SBML-Spatial, PEtab-MS, https://gitlab.com/fitmulticell/fit |
| Jürgen Pahle Heidelberg University | Oct 5 Oct 6 Oct 7 Oct 8 | - Copasi, CoRC |
| Lara Bruezière Novadiscovery | remotely | SBML;SED-ML, OMEX, COMBINE;SBGN Jinko software - collaborative clinical trial simulation platform |
| Lea Seep University Bonn, IRU-MLS | Oct 5 Oct 6 Oct 7 Oct 8 | SBML;Ontologies (SBO, KiSAO) - |
| Leslie Loew U. Conn. School of Medicine | Oct 5 Oct 6 | SBML;SED-ML, OMEX, COMBINE;Multicellular modeling Virtual Cell (aka VCell); SpringSaLaD |
| Lucian Smith University of Washington | Oct 5 Oct 6 Oct 7 Oct 8 | SBML;SED-ML, OMEX, COMBINE;Ontologies (SBO, KiSAO);Multicellular modeling Antimony, Roadrunner, Tellurium, SBML, SED-ML |
| Luis Fonseca University of Florida | remotely | SBML;SBGN;CellML;NeuroML;Multicellular modeling;ABM - |
| Lukas Buecherl University of Colorado Boulder | remotely | SBML;SED-ML, OMEX, COMBINE;SBOL, SBOL visual - |
| Lutz Brusch Technische Universität Dresden, Germany | remotely | Multicellular modeling Multicellular modeling, https://MultiCellML.org, MorpheusML, https://morpheus.gitlab.io, SBML-Spatial, PEtab-MS, https://gitlab.com/fitmulticell/fit, FAIRSPACE |
| Maren Philipps University of Bonn | Oct 5 Oct 6 Oct 7 Oct 8 | - PEtab, pyPESTO |
| Mauro Silberberg Universidad de Buenos Aires, Facultad de Ciencias Exactas y Naturales, Departamento de Física, y CONICET - Instituto de Física de Buenos Aires (IFIBA). Buenos Aires, Argentina | remotely | SBML;SED-ML, OMEX, COMBINE;CellML;NeuroML poincaré (github.com/maurosilber/poincare) and SimBio (github.com/hgrecco/simbio), which are Python libraries for definition and simulation of dynamical systems. |
| Michael Blinov Center for Cell Analysis and Modeling, UConn Health, | Oct 5 Oct 6 Oct 7 Oct 8 | SBML;SED-ML, OMEX, COMBINE;SBGN;BioPAX;Ontologies (SBO, KiSAO);Multicellular modeling VCell, BNGLViz, MolClustPy, |
| Michael Getz PostDoc | Oct 5 Oct 6 Oct 7 Oct 8 | SBML;CellML;Multicellular modeling - |
| Mustafa Ozen Altos Labs | Oct 7 Oct 8 | SED-ML, OMEX, COMBINE;CellML;NeuroML;Multicellular modeling - |
| Natasa Miskov-Zivanov University of Pittsburgh | Oct 5 Oct 6 Oct 7 Oct 8 | SBML;SED-ML, OMEX, COMBINE;SBGN;BioPAX;SBOL, SBOL visual;CellML;NeuroML;Ontologies (SBO, KiSAO);Multicellular modeling - |
| Nilesh Kumar PhD | Oct 5 Oct 6 Oct 7 Oct 8 | SBML;SED-ML, OMEX, COMBINE;CellML - |
| Norma Perez Rosas Purdue University | remotely | SBML;SED-ML, OMEX, COMBINE;SBGN;Multicellular modeling I work with mathematical models (ordinary and partial differential equations, kinetic modeling) to explain calcium activity in different biological systems. |
| Olga Krebs Heidelberg Institute for Theoretical Studies HITS | Oct 5 Oct 6 Oct 7 Oct 8 | SED-ML, OMEX, COMBINE;SBOL, SBOL visual;Ontologies (SBO, KiSAO);BioProtocols FAIRDOM, LiSyM Cancer, MESI-STRAT, PoLiMeR, deNBI |
| Paola Vera-Licona UConn Health | Oct 5 Oct 6 Oct 7 | SBML;SED-ML, OMEX, COMBINE;CellML;Multicellular modeling NETISCE (NETwork-drIven analysiS of CEllular reprogramming) http://veraliconalab.org/Netisce/index.html |
| Paul Jonas Jost University of Bonn | Oct 5 Oct 6 Oct 7 Oct 8 | SBML;CellML;Multicellular modeling - |
| Pedro Cenci Dal Castel Indiana University Bloomington | Oct 8 | SBML - |
| Pedro Mendes University of Connecticut School of Medicine | Oct 5 Oct 6 Oct 7 Oct 8 | SBML;SED-ML, OMEX, COMBINE;Ontologies (SBO, KiSAO);Multicellular modeling - |
| Prashant Vaidyanathan Oxford Biomedica | remotely | SBOL, SBOL visual - |
| Rahuman Sheriff European Bioinformatics Institute (EMBL-EBI) | Oct 6 Oct 7 Oct 8 | SBML;SED-ML, OMEX, COMBINE;SBGN;Ontologies (SBO, KiSAO);Multicellular modeling;FROG BioModels https://www.ebi.ac.uk/biomodels |
| Sarah Keating University College London | remotely | SBML;SED-ML, OMEX, COMBINE - |
| Sebastien Moretti SIB Swiss Institute of Bioinformatics | remotely | SBML;Ontologies (SBO, KiSAO) - |
| Sikao Guo PostDoc | Oct 5 Oct 6 Oct 7 Oct 8 | SBML;BioPAX;CellML;NeuroML;Multicellular modeling I am currently working on developing and parallelizing NERDSS (Structure-Resolved Reaction-Diffusion Simulation Software). You can find the GitHub page at https://github.com/mjohn218/NERDSS. |
| Sven Sahle Heidelberg University | Oct 5 Oct 6 Oct 7 | SBML;SED-ML, OMEX, COMBINE COPASI |
| T.J. Sego University of Florida | Oct 5 Oct 6 Oct 7 Oct 8 | SBML;Multicellular modeling CompuCell3D, Tissue Forge, https://directory.ufhealth.org/sego-t-j |
Breakout sessions
SBOL Visual Prashant Vaidyanathanm (Oxford Biomedica)
In these breakout sessions, we will develop into two exciting topics. First, we will explore the SBOL Visual Gallery, a novel initiative aimed at showcasing the diverse applications of SBOL visual diagrams. We will discuss strategies for encouraging the community to share their SBOL visual images and consider ways to link these images to SBOL examples. Additionally, we will brainstorm ideas for promoting these images to a wider audience through social media platforms such as Twitter and LinkedIn. Second, we will commemorate the 10th anniversary of SBOL visual by reflecting on its journey and impact over the past decade. We will discuss the evolution of SBOL visual, its contributions to the field, and its future prospects. Join us for a lively discussion on the past, present, and future of SBOL visual.
SBOL Outreach Prashant Vaidyanathan (Oxford Biomedica) In these breakout sessions, we will address several key topics related to the SBOL community. First, we will discuss the creation of an SBOL Projects portal or dashboard, a new effort designed to highlight tasks and ongoing projects that the community can contribute to. Second, we will explore the SBOL Release cadence, discussing strategies to ensure that the community can benefit from the latest version of SBOL. We will consider ways to keep our tools and models up-to-date to maximize the benefits for the community. Third, we will discuss outreach efforts, exploring ways to reach a wider SBOL audience, increase accessibility, and boost engagement. Lastly, we will discuss SBOL's involvement with the iGEM community and participants, brainstorming ways to enhance our collaboration and support. Join us for a productive conversation on how to strengthen the SBOL community and increase its impact.
SED2: The next generation of Simulation Experiment Design Eran Agmon, Lucian Smith, UConn Health
Intended Audience: Researchers, bioinformaticians, software developers, and modelers keen on shaping the future of in silico experiment design standards. Familiarity with SED-ML (https://sed-ml.org/) is advantageous but not essential.
Overview: The evolution of computational biology demands a more adaptable and integrative exchange format. While SED-ML has been instrumental, the emerging needs of the community call for a successor: SED2. This session will focus on the core priorities for SED2: flexibility, the ability to compose different simulation methods with custom annotations, and fostering a computational framework that works with the diverse range of simulation tools within the biological modeling community.
Session Goals:
- SED-ML Reflection: A recap of SED-ML’s journey, emphasizing its strengths and areas of improvement.
- Core Priorities for SED2: Deep dive into the need for flexibility and compositionality, and the significance of custom annotations.
- Building a Collaborative Framework: Engage in discussions to establish a common ground where members of the Biosimulations community can converge on SED2’s direction.
- Exchange Format and API Exploration: Deliberate on the potential of adopting formats like JSON for SED2 and brainstorm on the design of an intuitive API for seamless interactions with the new format.
Standardizing Multicellular Simulation - Bridging Methods and Models Eran Agmon (UConn Health), T.J. Sego (U of Florida); James Glazier (Indiana University)
Intended Audience: Researchers, bioinformaticians, software developers, and modelers who are involved in or interested in multicellular simulations across various methodologies. Prior experience with basic concepts in computational biology or multicellular modeling is beneficial but not mandatory.
Overview: Multicellular simulations have become indispensable in understanding complex biological phenomena, from tissue development to disease progression. But the diversity in simulation methods ‚ from agent-based models, cellular Potts models, cellular automata, lattice-free models, stochastic particle simulations, etc‚ poses challenges in reproducibility, modularity, reusability, and integration within multi-scale simulation. This session aims to bridge these gaps by focusing on the development of standards and schemas, with special emphasis on multiscale, embedded, and coupled simulation methods. Through a combination of presentations, case studies, and discussions, attendees will gain an understanding of the multicellular simulation landscape, the need for standardization, and the importance of sharing and reusing models.
Session Goals:
- Landscape of Multicellular Simulations: An overview of the various methods employed in multicellular simulations, highlighting their unique features and common challenges.
- The Need for Standards: Discussion on the current gaps and inconsistencies in multicellular simulation methodologies and the implications for research reproducibility and collaboration.
- Designing Schemas for Multicellularity: Collaborative brainstorming on creating robust and flexible schemas that can cater to the diverse range of multicellular simulation methods.
- Integration and Interoperability: Delve into strategies for ensuring that the developed standards promote seamless integration and interoperability among different simulation tools and platforms.
FROG curation for genome-scale metabolic models Karthik Raman, Rahuman Sheriff and other FROG contributors (Indian Institute of Technology, Madras)
Community standards for consistent reconstruction, FAIR sharing and curation of constraint-based models such as genome-scale metabolic models (GEMs) are crucial to ensure their reproducibility and reliability. We initiated a community effort for a standardised assessment of reproducibility and curation of constraint-based models. Following the discussions at dedicated breakout sessions at HARMONY and COMBINE meetings over the past two years, we have developed the FROG analysis, an ensemble of analysis of constraint-based models to test the reproducibility of numerical simulation based on a set of standardized analyses. FROG analysis encompasses Flux variability analysis (FVA), Reaction deletion analysis, Objective function calculation, and Gene deletion analysis. We have also developed a collection of tools that generate FROG reports in a standardized schema to enable a reliable assessment of model reproducibility based on popular constraint-based software and a web-service. FROG analysis is currently used in BioModels’ workflow to curate and build a collection of FAIR and reproducible genome-scale metabolic models.
Recent Progress and Future Work Towards Reproducible Stochastic Biological Simulation T.J. Sego (University of Florida); Rahuman S. Malik Sheriff (European Molecular Biology Laboratory)
Stochastic simulations are commonly used to quantitatively or semi-quantitatively describe the dynamics of biological systems. At various scales and in multiple applications, stochastic simulation better reflects observed biological processes and robustness. Various methods are widely used to incorporate stochasticity into biological simulation, such as the Gillespie stochastic simulation algorithm for systems biology modeling, stochastic Boolean networks for network modeling, and the Cellular Potts model methodology for multicellular modeling. Proving reproducibility of simulation results is critical to establishing the credibility of a model. To this end, BioModels, the largest repository of curated mathematical models, tests and reports the reproducibility of simulation results for all submitted models when possible. A recent study showed that about 50% of the deterministic ordinary differential equation models on BioModels could not be reproduced when applying criteria for reproducibility to the information provided in their associated publication, reflecting a current crisis of reproducibility. Furthermore, there are no well-accepted metrics or standards for reproducing stochastic simulation results, thus perpetuating the crisis of reproducibility for a broad class of biological models. This breakout session aims to establish an accepted framework for testing the reproducibility of stochastic simulations in biological modeling. The session will provide an overview of recent progress towards defining quantitative measures to determine whether stochastic simulation results can be reproduced, and when results have been reproduced. Attendees will discuss current issues to address towards consensus and broad adoption in relevant modeling communities, as well as future work towards reproducibility of stochastic simulation results using multiscale and complex models.
PEtab: current state and future directions Dilan Pathirana, Frank Bergmann (University of Bonn)
PEtab is a standardized file format for specifying parameter estimation problems [1]. The interoperable format is currently supported by 11 different tools [2], enabling users to benefit from standardized parameter estimation across frameworks based in Python, Julia, R, MATLAB, C++, or GUIs.
Although PEtab was initially developed for parameter estimation, recent efforts have extended the format to improve standardization of various adjacent tasks, including: model selection, multi-scale modeling, PKPD and NLME modeling, optimal control, and visualization.
In this breakout session, based on audience interests, we will present introductions to PEtab and its extensions, then discuss current efforts to improve PEtab. People unfamiliar with PEtab are welcome to attend, and might first like to check out the tutorial [3].
[1] “PEtab‚ Interoperable specification of parameter estimation problems in systems biology” https://doi.org/10.1371/journal.pcbi.1008646 [2] https://github.com/PEtab-dev/petab#petab-support-in-systems-biology-tools [3] https://petab.readthedocs.io/en/latest/tutorial.html
Annotations for SBML-qual models John Gennari (University of Washington)
Annotations against standard ontological resources is an important step for model reuse, model merging, and model comprehension. What are the specific needs for annotation of “logical” models (sometimes known as Boolean models)? In this breakout session, we’ll look at some example SBML-Qual models, especially gene regulatory networks. The specific augmentations that SBML-qual provides over “plain” SBML means that there are new opportunities to provide specific types of annotations on elements. As with any annotation effort, we will discuss tool support and ways to make annotation semi-automatic or easier for the modeler. We will also discuss how in-line annotations might look within SBML-qual, versus a separate file (per the COMBINE community recommendation).
A COMBINE Standard for Digital Twins of Living System?Rahuman Sheriff (European Bioinformatics Institute, EMBL-EBI)
Digital Twins are highly accurate and dynamic virtual replicas of real-world systems that has revolutionized a wide range of engineering industry. However, as this paradigm shift extends its reach into the realm of healthcare and medicine, including the ambitious endeavour of creating a human digital twin, it becomes evident that a standardized approach is imperative.
The COmputational Modeling in Biology Network (COMBINE) community, renowned for its development of critical standards such as SBML, CellML, SEDML, and PETab, now faces a pivotal question: Can it deliver a standardized specification for Digital Twins of patients, encompassing the complexities of human biology and beyond?
In this engaging breakout session, we will explore this compelling proposition. Together, we will delve into the pressing need for a COMBINE standard tailored to the model human Digital Twins. In this session, we will engage in discussion, debating the feasibility and implications of developing a universal standard for Digital Twins that promises to shape the future of personalised medicine. If the outcome of the discussion is positive, we could consider writing a white paper on this topic.
Tutorials
Biological and Biophysics Simulation in Tissue Forge: Introduction and Guided Simulation Building T.J. Sego (University of Florida)
Tissue Forge is open-source simulation software for interactive particle-based physics, chemistry and biology modeling and simulation. Tissue Forge allows users to create, simulate and explore models and virtual experiments based on soft condensed matter physics at multiple scales, from the molecular to the multicellular, using a simple interface. While Tissue Forge is designed to simplify solving problems in complex subcellular, cellular and tissue biophysics, it supports applications ranging from classic molecular dynamics to agent-based multicellular systems with dynamic populations. Tissue Forge users can build and interact with models and simulations in real-time and change simulation details during execution, or execute simulations off-screen and/or remotely in high-performance computing environments. Tissue Forge provides a growing library of built-in model components along with support for user-specified models during the development and application of custom, agent-based models. Tissue Forge includes an extensive Python API for model and simulation specification via Python scripts, an IPython console and a Jupyter Notebook, as well as C and C++ APIs for integrated applications with other software tools. Tissue Forge supports installations on Windows, Linux and MacOS systems and is available for local installation via conda. This workshop introduces the basic concepts, modeling and simulation features, and some relevant modeling applications of Tissue Forge through guided simulation scripting. Workshop concepts will introduce basic Tissue Forge modeling concepts and simulation features through the development of interactive simulations in Python. Attendees are encouraged, but not required, to code along as the workshop interactively develops and tests simulations in multicellular and biophysics modeling applications.
Bridging the Gap - A Practical Guide to Model Specification Translation in Agent-Based Modeling Juliano Ferrari Gianlupi (UTHSC)
In the world of Agent-Based Modeling (ABM), the quest for cross-platform portability and model reproducibility is a formidable challenge. This tutorial is designed to empower modelers, scientists, and researchers with the knowledge and skills to overcome this challenge using a novel Model Specification Translator.
Agent-Based Modeling has emerged as a vital tool for exploring intricate biological systems, from cancer progression to embryonic development. However, the lack of interoperability and reusability among ABM platforms has raised concerns about model reproducibility. Our tutorial addresses these concerns head-on, introducing a practical and hands-on approach to translate models across different platforms.
During this tutorial, participants will embark on a step-by-step journey through the Model Specification Translation process. We will dive into the essential concepts and methodologies necessary for seamlessly converting models from one platform to another. Whether you’re working with CompuCell3D, Tissue Forge, PhysiCell, or any other ABM platform, this tutorial will equip you with the skills needed to ensure your models remain portable and interoperable.
Additionally, we will showcase PhenoCellPy, a Python package that simplifies the creation of cell behavioral patterns. This tool not only enhances model accessibility for biologists but also streamlines the transition from biological concepts to computational implementation.
Participants should have python, a git interface (optional), and CompuCell3D (https://compucell3d.org/SrcBin) installed. I suggest installing CC3D using conda. Please also install the xmltodict python package (https://pypi.org/project/xmltodict/).
Join us in this tutorial to explore the significance of cross-platform portability, learn how to overcome the intricacies of model porting, and contribute to the broader ABM community’s effort in establishing a universal modeling description standard. By the end of this tutorial, you will be better equipped to advance agent-based modeling, foster model reproducibility, and gain deeper insights into complex biological systems (https://github.com/JulianoGianlupi/pcxml2cc3d).
Mastering Structure-Resolved Reaction-Diffusion Simulations with NERDSS Sikao Guo (Johns Hopkins University)
This tutorial (https://sikaoguo22.github.io/NERDSSTutorial/) is intended for researchers, students, and professionals in cellular biology, biophysics, and computational biology who are interested in spatiotemporal reaction-diffusion simulation. NERDSS (https://github.com/mjohn218/NERDSS) is a nonequilibrium reaction-diffusion self-assembly simulator that integrates molecular structures and their processes to understand the dynamics of cellular processes that last for minutes (https://doi.org/10.1016/j.bpj.2020.05.002). It allows users to build a reaction-diffusion model based on actual molecular structures, which enhances the model’s accuracy and captures the complexities of multisubunit complexes and their reversible formation. Several case studies have employed NERDSS, such as the formation and spontaneous disassembly of large clathrin lattices (https://doi.org/10.1371/journal.pcbi.1009969), the dynamic behavior of the HIV Gag lattice in virions (https://doi.org/10.7554/eLife.84881), and understanding the temporal influence of cofactors in retroviral Gag lattice assembly (https://doi.org/10.1016/j.bpj.2023.06.021). During the session, we will explore these applications, demonstrating the software’s versatility in handling diverse cellular processes. The session will cover core principles of structure-resolved reaction-diffusion, emphasizing its role in cellular biology. We will attempt to build a coarse-grained structure from the real protein structure from the pdb database. Then, we will learn how to set up a model, run the model with NERDSS, analyze and visualize the model outcomes with io_nerdss (https://github.com/mjohn218/io_nerdss) and OVITO.
Tutorial on biological modeling with PySB Mustafa Ozen Ryan Spangler, Carlos F. Lopez (Altos Labs)
PySB (Python Systems Biology) is a powerful and versatile biological modeling tool that has gained prominence in systems biology. It provides a Python-based programming platform with a rule-based framework for constructing dynamic models of sophisticated biochemical systems, enabling researchers to simulate and analyze complex cellular processes. This unique tool empowers researchers to modularly define molecular interactions and transformations, facilitating the representation of a wide array of biological processes in a simple, interpretable way. PySB accommodates both stochastic and deterministic simulation methods, providing a comprehensive view of system behavior. Its seamless integration with various model calibration, analysis, and visualization libraries further assists researchers in interpreting simulation results effectively. In this tutorial session, we aim to walk the attendants through the foundations of PySB, show them how it works, and provide them with hands-on experience.
Relevant Resources: PySB Paper: https://www.embopress.org/doi/full/10.1038/msb.2013.1 PySB Website: https://pysb.org/ PySB GitHub: https://github.com/pysb/pysb PySB Tutorial: https://pysb.readthedocs.io/en/stable/tutorial.html
Introducing poincaré and SimBio Mauro Silberberg (Universidad de Buenos Aires, Facultad de Ciencias Exactas y Naturales, Departamento de Fisica)
Poincaré is a new Python-based library to define and simulate dynamical systems. Trying not to reinvent (much of) the wheel, it uses modern Python syntax to define and compose models in a compact way. This allows us to take advantage of the huge investment on tooling from the broader Python community such as static type checkers, code linters and formatters, autocomplete and refactoring features from IDEs, among others.
Using standard libraries from the PyData ecosystem, by default, poincare compiles into a first-order ODE system using NumPy arrays, and uses solvers from SciPy. But it also provides different backends such as Numba, which compiles just-in-time to LLVM code, providing a significant speed boost, or JAX, which provides autodifferentiation tools targeted for ML. Additionally, it supports units using the Pint library.
SimBio is built on top of poincaré, adding some components for reaction-based models used in systems biology. It provides some predefined building blocks for the most common reactions, but allows easily to create your own. Finally, it implements an importer and exporter to SBML, allowing to interexchange models with the COMBINE community. We hope that it is simple enough for beginners, but powerful for power-users with the possibility to extend and compose with the large Python ecosystem.
DySE: Dynamic System Explanation framework
Difei Tang (University of Pittsbrugh); Natasa Miskov-Zivanov (University of Pittsburgh)
In this demo, we will showcase our framework, DySE (Dynamic System Explanation), that includes tools for model simulation, model extension, interaction classification, interaction filtering, model checking, and sensitivity analysis. The rapid proliferation of data generated by experiments studying biological systems poses a considerable challenge. This overflow of information is spread across an array of publishing platforms, making it increasingly difficult to manually analyze all available data. This underscores the necessity for automated methods that can retrieve and connect relevant pieces of this voluminous knowledge. Such methods are crucial for understanding, explaining, and predicting the behavior of these complex systems.
To tackle this challenge, DySE integrates machine reading, automated model assembly, and computational analysis to enhance understanding and explanation of complex systems. The toolset is conveniently accessible via a user-friendly graphical interface (GUI).
FLUTE utilizes existing databases to evaluate confidence and trustworthiness of given biochemical interactions (https://tinyurl.com/flutedoc). VIOLIN classifies large sets of interactions with respect to a given model. The interactions are classified into four main categories, corroborations, contradictions, extensions, and flagged, and several subcategories within main ones (https://tinyurl.com/violindoc). CLARINET (see https://tinyurl.com/clarinetdoc and link to binder notebook on main page) and ACCORDION (see https://tinyurl.com/accordiondoc and link to binder notebook on main page) automatically expand and recommend models based on selected desired model properties.
In conjunction with these front-end tools, we’ve also developed back-end tools within the DySE framework. DiSH is a stochastic simulator offering versatile simulation schemes and timing options (https://tinyurl.com/dishjupyter). PIANO provides comprehensive sensitivity analysis for the entire model, and allows for identifying most influential pathways and suggesting interventions (https://tinyurl.com/pianojupyter). Additionally, we’ve introduced a unified format compatible with the tools mentioned above: BioRECIPE (https://tinyurl.com/biorecipe), seamlessly translating to and from widely used synthetic biology modeling languages.
All of these tools can be used either independently or in combination. For instance, synthetic biologists can rely on it to ensure the reliability of interactions in designing synthetic biological systems. Bioinformaticians can efficiently filter and prioritize data, while computational modelers benefit from model extensions to always get model up-to-date. Professionals in biotech industries utilize sensitivity analysis for optimizing therapies. Additionally, even educators would find it valuable when using simulation software.
Novel advances in the automation of knowledge selection and model assembly Natasa Miskov-Zivanov, Yasmine Ahmed, Gaoxiang Zhou (University of Pittsburgh)
Creating computational models of complicated systems, including intracellular and intercellular bionetworks, is a time and labor-intensive task which is often limited by the knowledge and experience of Pathway database modelers. This has naturally led to the emergence of the idea of automating the process of building new/extending existing models, which could have a significant potential in enabling rapid, consistent, comprehensive and robust analysis of complicated systems. Inspired by this idea, we propose in this work different novel approaches namely ACCORDION (ACCelerating and Optimizing model RecommenDatIONs) and CLARINET (CLARifying NETworks) for expanding models using the information extracted from literature by machine reading engines. Our proposed approaches combine machine reading with clustering, and graph theoretical analysis to create an automated framework for efficient model assembly. Furthermore, by automatically extending models with the information published in literature, our proposed methods allow for collecting the existing information in a consistent and comprehensive way. This, in turn, facilitates information reuse, data reproducibility, and replacing hundreds/thousands of manual experiments, thereby reducing the time needed for the advancement of knowledge. To evaluate ACCORDION1 and CLARINET, we compare their outcomes with three previously published manually created models namely naive T cell differentiation model, T cell large granular lymphocyte leukemia model and pancreatic cancer cell model. Besides demonstrating automated reconstruction of a model that was previously built manually, our tools can assemble multiple models that satisfy desired system properties. As such, they replace large number of tedious or even impractical manual experiments and guide alternative hypotheses and interventions in biological systems.
A GitHub page, ReadtheDocs and Jupyter notebook are available for ACCORDION http://www.nmzlab.pitt.edu/accordion and CLARINET http://www.nmzlab.pitt.edu/clarinet.
Using Compucell3D as a Platform for Model Building to Explore Cell Behaviors, Cell-Cell Interactions, Cell Migration and Chemotaxis Pedro Dal-Castel (Biocomplexity Institute and Department of Intelligent Systems Engineering, Indiana University)
Mechanistic agent-based modeling is an integral part of contemporary bioscience, used for hypothesis generation and testing, experiment design and interpretation, and the design of therapeutic interventions. The CompuCell3D (CC3D) modeling environment allows researchers to rapidly build and execute complex virtual tissue simulations with minimal programming experience. CC3D enables biological simulations from subcellular to tissue scales, supporting explicit cell shapes, cell migration, contact-mediated cell interactions, soluble signals, and complex cell state dynamics (gene regulatory, signaling, and metabolic networks). CC3D natively supports SBML, Antimony, and MaBoSS network model integration. Participants will (1) learn how to build models in CC3D, (2) implement network models in CC3D, and (3) develop an example simulation with all concepts learned. CC3D can be accessed from the official website (www.compucell3d.org) or running it on-line at (https://nanohub.org/tools/cc3dbase4x ).
Audience: Anyone interested in multicellular Virtual-Tissue modeling or in coupling network models to cell behaviors and dynamic spatial organization. If you plan on participating, please fill out this form: https://forms.gle/TTnw88hR3PebCkmE7
Learning Outcomes: Ability to use CompuCell3D to design, execute and explore virtual-tissue simulations integrating cells, networks and external chemical fields.
Computer Requirements: Any Windows or Mac computer. CompuCell3D is open-source and free. It also runs on many LINUX deployments (see www.compucell3d.org for details). Our preferred method for this miniworkshop is launching CC3D from nanoHUB. We ask that you open an account in advance (https://nanohub.org/register/ ).
Class materials: https://drive.google.com/drive/folders/1RJMgfO9PDGvpfJ5Fp7OvVouNK9DvDuW5?usp=drive_link Zoom session https://iu.zoom.us/j/4555920347 For more information, see https://www.compucell3d.org or contact pdalcastel@gmail.com, or jaglazier@gmail.com
Talks
Multicellular and Multiscale Models of Microbes and Host Systems Elebeoba May (U Wisconsin-Madison)
Computational and experimental models enable concurrent spatiotemporal and biochemical modeling, monitoring and characterization of microbe-microbe and microbe-host interactions to understand bacterial communities and infection dynamics and outcome. We have developed multiple in silico intracellular and multicellular models to probe the contribution of physiological, structural and biochemical processes in control, resolution or dissemination of bacteria in models of bacterial communities and mycobacterium infection. In this talk, we will highlight multiscale models including models of Mycobacterium tuberculosis (Mtb), an intracellular pathogen, that can adapt to changing environments within host phagocytic cells enabling persistence and proliferation during infection and onset of disease. Our models integrate host and pathogen interactions using a multiscale, agent-based models (ABM). Models are informed by empirical characterization studies and account for changes in the physiological environment of the host as well as the host intracellular environment. We discuss expanded models and challenges in developing models to explore response in individuals with chronic obstructive pulmonary disease (COPD) and tuberculosis.
Multi-Scale Multicellular Agent-Based Virtual-Tissue Simulations: Challenges and Opportunities in Sharable Model Specification
James Glazier (Indiana University)
Multi-scale, Multicellular Agent-Based Virtual-Tissue models built using modeling frameworks like CompuCell3D are versatile tools for exploring the complex interactions between signaling and gene- regulatory networks, icell-cell signaling through contact and diffusible signals, and force generation, cell migration, proliferation and shape change, and finally for coupling to much higher scales such as the whole body, the environment and populations. They can play a crucial role in helping to interpret and design more informative experiments, such as in vitro to in vivo extrapolation. However, Virtual Tissues currently lack model-specification standards, support for modular architectures and annotation, cross-compatible tools for model specification, visualization and analysis, and a model sharing infrastructure. This last aspect of a model sharing infrastructure has enabled the rapid development of systems biology network modeling as a core technology in research and is gaining wider acceptance in regulatory environments. Comparable infrastructure is essential for Virtual Tissues to move from academic one-offs for basic research to mainstream technologies in biomedicine, technology and regulation. Because Virtual Tissues are substantially more complex and structurally and functionally more diverse than network models, standardization and modularization, graphical specification and distribution are all more challenging. I will consider some of the variety of Virtual Tissue applications, frameworks and modeling approaches and some of the challenges and opportunities we face in developing an effective ecosystem of tools and standards.
Breaking Barriers in Multiscale Agent-Based Models: A Path for Cross-Platform Models Juliano Ferrari Gianlupi (UTHSC)
Agent-based modeling (ABM) is a powerful tool for understanding complex biological systems such as cancer progression, embryonic development, and pathogen infections. Ensuring reliable and accurate simulations is crucial as ABMs gain prominence. This research focuses on addressing the crisis of reproducibility in ABMs through two approaches. First, we introduce a model specification translator to facilitate cross-platform model reproducibility. Second, we present PhenoCellPy, a cross-platform Python package that simplifies the creation of cell behavioral patterns, making modeling more accessible to biologists and improving inter-platform operability. Biological systems often exhibit stereotyped transitions between distinct phases, necessitating structured definitions of states and transitions. Translating cellular behaviors into computational models requires a deep understanding of both biology and modeling. To enhance model sharing and accessibility we created PhenoCellPy. We aim to create pattern specifications usable across various modeling platforms, streamlining the transition from biological concepts to computational implementation, as patterns would have to be recreated for each new model. The need for cross-platform portability and interoperability is critical for model validation. While challenges exist in achieving model portability, we emphasize the importance of establishing a universal modeling description standard akin to SBML, calling for a collaborative effort within the AB modeling community. We investigate some of the challenges this universal ABM specification will face, and make headway into its creation with our model specification translator. It can translate PhysiCell models into CompuCell3D models.
MolClustPy: a Python package to characterize multivalent biomolecular clusters Aniruddha Chattaraj (Harvard U)
Low-affinity interactions among multivalent biomolecules may lead to the formation of molecular complexes that undergo phase transitions to become supply-limited large clusters. In stochastic simulations, such clusters display a wide range of sizes and compositions. We have developed a Python package, MolClustPy, which performs multiple stochastic simulation runs using NFsim (Network-Free stochastic simulator); MolClustPy characterizes and visualizes the distribution of cluster sizes, molecular composition, and bonds across molecular clusters. The statistical analysis offered by MolClustPy is readily applicable to other stochastic simulation software, such as SpringSaLaD and ReaDDy.
FAIRDOM-SEEK: FAIR and standartised management of models and related (meta)data. Olga Krebs (Heidelberg Institute for Theoretical Studies, Germany)
FAIRDOM-SEEK (https://fairdomseek.org/) is an open source software for storing, cataloguing, sharing and reusing research outcomes, it is designed to support the principles of FAIR (Findable, Accessible, Interoperable, and Reusable) research data management. FAIRDOM-SEEK offers features that enable scientists to organize, document, and publish their research data. The ISA (Investigation, Study, Assay) standard framework supports the organization and description of individual experiments and related research items, such standard operating procedures, or protocols. Key components of FAIRDOM-SEEK include data and metadata management, version control of files, and integration with modelling tools, e.g. JWS online and Copasi. Additionally, FAIRDOM-SEEK promotes collaboration by providing mechanisms for sharing and exchanging metadata and research outputs with specific individuals, research groups, and the public. The possibility to create persistent identifiers (DOI) for public data files or experiments allows for long term access to research outputs that can be used as e.g. supplementary material in publications. The SEEK platform enables the building of Project Hubs where investigators can store, share, access, connect and interact with digital objects generated from their research, and use them in their own analyses. FAIRDOMHub is used as a research data management platform by 350 national and international collaborative projects not only as a data repository but also as a knowledge-sharing platform. It encourages the creation of communities and fosters interdisciplinary collaborations by connecting researchers with shared interests and expertise. FAIRDOMHub is developed and maintained by the FAIRDOM (https://fair- dom.org/) consortium and is a key component of the data management services of de.NBI, the German Network for Bioinformatics Infrastructure (https://www.denbi.de/), and ELIXIR, the European Infrastructure for Life Sciences (https://elixir-europe.org/).
Simulation-Driven Evaluation of Genetic Circuit Architectures Using Combinatorial Designs with the Synthetic Biology Open Language (SBOL) Helen Scott (Raytheon BBN)
The field of synthetic biology has advanced by developing “parts”, genetic elements and tools with defined functions, and then combining these parts to create circuits with increasing complexity. As DNA synthesis technology and assembly methods have improved, synthetic biologists are now able to combine these parts into more complex circuits with more advanced functions. However, when designing a circuit, fully exploring the combinatorial space of relevant parts can be time and cost prohibitive to carry out in the lab. One possible way to exhaustively search this combinatorial space is to construct a SBOL generator for each modular element of the genetic circuit. These elements can then be assembled combinatorically to generate a genetic regulatory network for each possible configuration. An SBOL representation of these circuits can then aide in simulation-based engineering, as LaTeX equations and MATLAB code can be generated programmatically for each such genetic regulatory network. Here we present two uses of combinatorial design with SBOL for the comparative evaluation of potential genetic circuit architectures and their sensitivity to parameter values: the first modularly combined regulatory elements to investigate time- delayed safety switches for CRISPR gene therapies, and the second combined CRISPRi-gRNA target sites to investigate architectures for synthetic plant promoters. We anticipate that by aiding in combinatorial design, SBOL can allow for faster engineering of genetic circuits across many applications. This work was supported by DARPA contract HR0011-18-2-0049. This document does not contain technology or technical data controlled under either U.S. International Traffic in Arms Regulation or U.S. Export Administration Regulations. Views, opinions, and/or findings expressed are those of the author(s) and should not be interpreted as representing the official views or policies of the Department of Defense or the U.S. Government. Approved for public release, distribution unlimited (DISTAR Case 38663).
Knowledge-Based Pathway Extraction and Validation in Cell Signaling Networks Gaoxiang Zhou (UPitt) In system biology, achieving precise control in cell signaling networks is paramount, and pathway analysis is a vital tool in this endeavor. It helps identify key genes, enzymes, and reactions within pathways, offering insights into their interactions and behaviors. Nonetheless, current approaches primarily centered on establishing connections between isolated interactions of two spices frequently fall short in providing the essential biological context required for precise analysis. To tackle this issue, this study introduces a knowledge-based (KB) pathway extraction method and develops a hands-on python-based toolkit. By incorporating this approach and a rigorous validation scheme, system biologists gain a powerful method for precise pathway analysis. The methodology underlying this approach entails the assignment of scores to pathways and extraction based on path scores for certain source-target node pairs. Subsequently, a validation scheme is introduced to validate the extraction results. This scheme is designed to preserve the biological properties. Specifically, it aims to demonstrate that the removal of crucial pathways, achieved computationally through regulatory knockdown, should have an impact on these properties, while the removal of less significant pathways should not. To achieve this, a set of properties inherent to the original modeled system is established based on expert knowledge. Partial models are then created by eliminating the highest-ranked pathways. The evaluation of these partial models against the defined properties is conducted using a combination of statistical model checking and element-based simulations. The core of the methodology lies in the graph search algorithm, which identifies the most "important" paths in a weighted directed graph from a source to a target. Different interpretations of importance lead to diverse edge weight assignments. Nine attributes are proposed for weight assignment in this work, including in-degree, out-degree, shortest link, loop count, non-bias, immediate edge influence, and element sensitivity. Each attribute offers a unique perspective on pathway importance. They are elaborately compared and contrasted in the context of a case study. Our case study centers on a T-cell differentiation model, wherein we investigate various scenarios and evaluate the consequences of pathway removal based on diverse interpretations of significance. The findings illustrate that, in most scenarios, the model's behavior is most significantly affected when pathways are removed based on immediate edge influence and element sensitivity attributes. Immediate edge influence and element sensitivity are conceptualized to capture aspects of network topology, dynamics, and, notably, knowledge semantics. These outcomes underscore the critical role of knowledge-based pathway analysis in the study of biological behaviors.
Lightning talks
The Reproducibility Portal Herbert Sauro, Lucian Smith, Joe Hellerstein (U Washington)
Reproducibility is a foundation of science. In Systems Biology, this requires access to details of models and modeling results. Much of this information exists in websites such as BioModels and Biosimulations, but accessing the information can be complicated. The Reproducibility Portal (https://www.reproducibilityportal.org/) provides simple and fast access to information such as: the article summary, human-readable rendering of SBML models, and the ability to explore simulation runs. The Portal is a website hosted as a GitHub page.
Constrained Fitting of Rule-based Models with Simmune Fengkai Zhang (NIH)
Antimony at 15 Lucian Smith (U Washington)
Antimony is a human-readable, human-writeable model format, adapted from Jarnac in 2008 that can be translated to and from SBML. This has formed a lynchpin of our Tellurium system, enabling modelers to easily create models. Here, I cover more recent updates to the language and particularly the challenges of trying to easily annotate models.
SBML tools and browser based modelingb> Bartholomew Jardine (U Washington) REMOTE
Browser based, client-side applications are increasing in popularity as they have certain advantages over traditional native code and server-side applications. As they are browser based the user does not need to install it, just click the link and the application loads, and current browser javascript interpreters are highly optimized and execute javascript and web assembly at near native application speed. As a client-side application, the code is run on the user’s computer with no server-side needs which enables ‘dumb’ websites to host the application as the site is basically used as a file server. For the SBML (sbml.org) community we have two libraries: libsbmljs (https://github.com/sys-bio/libsbmljs) and libantimonyjs (https://github.com/sys-bio/libantimonyjs) and a couple of implementations: MakeSBML (https://sys-bio.github.io/makesbml/), an application that makes converting SBML models to Antimony easy and miniSidewinder (https://github.com/sys- bio/miniSidewinder), a lightweight simulator of SBML models, that use the libantimonyjs and libsbmljs libraries and show what is possible in browser based modeling and simulation.
Retroactive Curation: Lessons and Challenges Lucian Smith (U Washington)
Since 2005, curators at the EBI (European Bioinformatics Institute) have been curating papers, creating SBML models that can be used to reproduce figures in papers. With the advent of SED-ML (the Simulation Experiment Description Markup Language), it became possible to not only encode the model used to produce the figures, but also to encode how to create the figure from the model. At the Center for Reproducible Biomedical Modeling, we have been working on re-curating Biomodels to add SED-ML files, and run the resulting OMEX files at biosimulations.org on multiple simulators to confirm validity. To date, 829 SED-ML files run and produce the same results on multiple simulators, and 1035 SED- ML files run on at least one simulator.
ENSEMBLE MODELING OF BACTERIAL RESPONSE TO IRON AND OXIDATIVE STRESS Daniel Ajuzie (U Houston)
BACKGROUND: Iron regulation in bacteria is complex and influenced by environmental factors such as oxidants [1]. Models exist for cellular iron regulation and oxidative stress response, but few address the challenge of modeling multiple phenotypes. Our previous in-silico model integrated iron and hydrogen peroxide stress response in Escherichia coli K12. Here, we present a multi-phenotype approach for parametrizing the iron stress response model, to improve prediction accuracy across multiple phenotypes. METHODS: Experimental models have demonstrated that Escherichia coli (E. coli) can exhibit different phenotypes, including proliferative, bacteriostatic, or bactericidal, depending on the combination and intensity of stress factors. We integrated and expanded on previous models to construct a two compartmental model of that captures iron[2] and oxidative stress[3] interactions and resolves response in E. coli to varying concentrations of ferric iron, ferrous iron and hydrogen peroxide stress at different scales of genetic regulation, metabolic response and physiological response. We employed augmented sensitivity analysis [4] to identify influential parameters for our model and utilized multi-phenotype assumption (MP) and sequential parameter estimation in the optimization approach. Ensemble modeling methods were used to aggregate single models to improve prediction accuracy. To validate our model, we compared its ability to replicate the same response to stress conditions as the experiments. Additionally, we compared our MP approach with standard single phenotype (SP) approaches. RESULTS: The MP model accurately recapitulated 70 - 80% of experimental response variables across 20 distinct stress-response categories compared to 65 – 75% obtained using SP approaches. Ensemble modeling was found to reduce modeling error by up to 11% across the different categories for both the MP and SP models. Using the MP model, we identified siderophore production, growth, and peroxide-dependent transcriptional regulation as the most critical sub-components of the iron homeostatic machinery in E. coli necessary for developing an adequate predictive model. Additionally, our models explained bacterial siderophore response in the context of dynamic iron and peroxide stress. Results seem to suggest that enterobactin is causally correlated with cellular growth parameters and likely confers cellular protection under peroxide stress independent of iron stress. Our model simulations uncover that low levels of peroxide stress in an iron-rich environment pose a challenging scenario for bacterial adaptation, resulting in a bacteriostatic phenotype. This difficulty can be attributed to the intricate interplay between genetic regulation and metabolic response under this specific combination and intensity of stress. CONCLUSIONS: Understanding variations in bacterial persistence resulting from differences in the maintenance of iron homeostatic processes could aid in the identification of potential therapeutic targets or novel therapeutic strategies for host-pathogen dynamics. An immediate future application for this model would be its integration into multiscale model of biofilm formation [5] developed in our laboratory to understand how iron metabolism and oxidative stress impacts processes involved in biofilm formation.
Curating models from Biomodels: Developing a workflow for creating OMEX files Jin Xu (U Washington)
I examined a selection of models in the Biomodels Database. For each model, I reproduced the published results using Tellurium. Once reproduced I manually created a standard OMEX file using SBML and SEDML. These exercises have allowed me to develop a workflow that we will use to develop an online platform to help users more easily curate models for biosimulations.org and www.reproducibilityportal.org.
Introducing the BioRECIPE Format: A Human-Centric Representation For Computational Modeling in Systems Biology Gaoxiang Zhou, Difei Tang, Natasa Miskov-Zivanov (U Pitt)
In the field of systems biology, widely employed representation formats for computational modeling, such as SBML, CellML, and BioPAX, typically rely on markup languages and prioritize computer-friendliness. However, these formats often lack accessibility for human modelers and experts when it comes to the tasks of model creation, verification, evaluation, curation, and expansion. There is a growing demand for a format that enables human users to intuitively preview and modify various aspects of models, including individual model elements, interactions between these elements, and even the attributes associated with these interactions. In this study, we introduce the BioRECIPE (Biological system Representation for Evaluation, Curation, Interoperability, Preserving, and Execution) format to address this demand. BioRECIPE is primarily a tabular format, ideally suited for models structured as directed graphs, while retaining machine-readability and compatibility with a range of model development and analysis tools. The BioRECIPE format serves a dual purpose, allowing the representation of two distinct formats: event-based lists of interactions and element-based models. In the event-based format, each row corresponds to a single interaction, with column headers aligning with interaction attribute names, categorizing attributes into elements, interactions, context, and provenance. On the other hand, the element-based model format assigns each row to a model element and supports the representation of a model’s static graph structure along with attributes necessary for dynamic analysis. Unlike the event-based format, multiple interactions can be combined within element update rules. This format accommodates various model representation schemes, ranging from less to more detailed, including static and dynamic parameters. Additionally, this format incorporates three attribute groups for defining rules concerning element state changes, namely element regulation, value, and timing. The BioRECIPE format is presently compatible with established representation formats like SBML and JSON, facilitating seamless translation between various output formats obtained from REACH, TRIPS, and INDRA. Furthermore, it serves as a standardized format within the DySE framework and is integrated into many analysis tools inside, including FLUTE, VIOLIN, CLARINET, ACCORDION, FIDDLE, and PIANO. The detailed documentation of this BioRECIPE format can be accessed at https://melody-biorecipe.readthedocs.io/
Alcuin: A cross-platform Qt/C++ portable library to create an embeddable widget with customizable features required for a network viewer/editor software tool. Adel Heydarabadipour & Herbert M. Sauro. (U Washington) REMOTE
Alcuin is an open-source cross-platform Qt/C++ portable library that provides developers with an embeddable widget with the required features for a network viewer/editor software tool. With minimal effort, a developer can embed this widget into their own software tool and provide their users with a user interface to view, build, and edit a network consisting of nodes and edges. Alcuin is shipped with the fundamental features of a network viewer/editor tool, including adding/removing nodes to its graphics scene, adding/removing edges (with optional arrowheads) between nodes, nesting one node into another node, modifying the properties of network elements (nodes, edges, and arrowheads), selecting, moving, copying, cutting, pasting, aligning, dragging and dropping network elements, zooming and panning on its graphics scene, and undoing and redoing the user’s actions. In addition, Alcuin is equipped with a plugin interface that lets developers use python scripts to customize it at their will to meet the specific requirements of their users. As a use case, we show how its plugin interface can be implemented to customize this widget into a tool for viewing, building, and editing biochemical reaction networks formatted using SBML. Using this customized tool, a user can create, load, save or edit biochemical reaction networks compatible with SBML as well as the SBML Layout and Render. The name Alcuin refers to the 8th century English scholar and teacher who was headmaster in 778 AD at the cathedral school of York, England and later became master of the palace school of Charlemagne at Aachen in 782 AD.
The source code of the project along with its binary installers for Windows and macOS platforms are available on its GitHub page, https://github.com/adelhpour/NetworkEditorGUI.
Posters
cOmicsArt - customisable Omics analysis reporting tool Paul Jonas Jost (U Bonn)
Modern researchers often face a daunting challenge when dealing with the vast amounts of data generated by omics technologies. Navigating this data typically demands either a proficiency in programming or the need to seek assistance, both of which can impose constraints on the research exploration process.
To address this issue, we introduce a web-based software tool developed in R. This tool serves as an installation-free graphical user interface, designed to offer an interactive and accessible means for initial data exploration. Upon uploading the data matrix and sample names, the platform offers a wide array of data-selection options, diverse preprocessing techniques, and essential preliminary analyses with interactive features. These include Principal Component Analyses (PCAs), Volcano Plots, heatmaps, basic differential expression analysis as well as enrichment analysis.
A key feature ensuring research reproducibility is the automatic generation of a detailed exploration report. Every alteration made to the raw data is meticulously documented. Additionally, any plots that users choose to save are seamlessly integrated into the report, complete with their corresponding settings. Moreover, for those interested in further manipulating the plots, an R code can be readily downloaded. This code empowers users to reproduce and customize the plots directly within the R environment. Alternatively, the plots can be effortlessly downloaded in various image formats.
This tool democratizes the data exploration process, making it accessible to researchers regardless of their programming background. Researchers can now explore and experiment with their omics data, undertaking crucial initial steps in their analysis independently. The tool is designed to empower experimentalists to engage with their data according to their specific interests, thereby reducing the burden associated with subsequent analysis.
Metadata Management: A Synchronous Metadata Recording Approach Lea Seep (UBonn)
In scientific collaboration metadata management is critical for data reproducibility. However, wet-lab researchers need to patch together diverse set of metadata standards for the different stages during the data lifecycle. Consequently, metadata is frequently gathered retrospective at point of publication, which can miss pre-publication collaboration and knowledge sharing opportunities. Our introduced metadata structure aligns with the data lifecycle for synchronous metadata recording, covering the planning to data collection for an experimental design and measurement combination. Its implementation within Excel offers several features to streamline the recording process, including automatic structure selection, metadata integrity checks and export options for various formats required by repositories. The design prioritizes the ease of complete data recording over structural robustness. We anticipate the deployment of centralized database for metadata records by providing a suitable ontology. The proposed metadata structure simplifies recording and structuring for wet-lab scientists, promoting practicality, and convenience in metadata management. This framework can accelerate scientific progress by enhancing collaboration and knowledge transfer throughout intermediate steps of data creation.
pyPESTO: PYthon Parameter EStimation Toolbox Maren Philipps (U Bonn)
pyPESTO is a widely applicable and highly customizable toolbox for parameter estimation. It can be used as a parameter estimation pipeline for systems biology problems specified in SBML and PEtab. The interface to AMICI enables efficient simulation and sensitivity analysis of ordinary differential equation (ODE) models. Toolbox features include multi-start local optimization, profile computation and visualization. Notably, it accommodates parameter estimation with ordinal data, censored data and nonlinear-monotone data.
Model selection with PETAB Dilan Pathirana (U Bonn)
Model selection is the task of identifying useful models from a set of plausible models. Often, this involves calibration of models, then subsequent comparison of models by some criterion, e.g. the Akaike (AIC) or Bayesian information criterion (BIC). PEtab itself is a file format to specify the parameter estimation problem for a model, including the objective function noise model and data, which facilitates calibration of single models. We extend PEtab to support model selection (PEtab Select), by (1) designing a concise file format for the specification of models and hyperparameters, and (2) creating a software library for model selection algorithms. This software library enables tools that already support the PEtab format, to easily add model selection capabilities, by simply interfacing the PEtab Select library. PEtab Select is currently supported by some popular systems biology tools, including COPASI, PEtab.jl, and pyPESTO. Implemented algorithms include stepwise methods like forward and backward, and well as brute- force and the state-of-the-art stepwise method FAMoS. PEtab Select has been used to solve large model selection problems from the literature, including a synthetic-data problem with 65,000 models, and a real- data problem with 4 billion models.
Time-dependent parameters and optimal control with PEtab Dilan Pathirana (U Bonn)
Models in systems biology can involve time-dependent parameters. These can be used to encode evolving governmental interventions strategies during a pandemic, or adjustable patient-specific drug regimens. The values that these parameters take over the timecourse are typically considered independent of the mathematical model that describes the dynamics of the system. Hence, keeping them separate from the model can help make patient-specific models more modular. PEtab is a file format to specify the parameter estimation problem for a model, such that the model can be calibrated. We extend PEtab to support complex experimental conditions involving time-dependent parameters (PEtab Timecourse). We further extend this to support optimal control of these time-dependent parameters (PEtab Control). In both extensions, we (1) introduce new file formats to concisely specify the new information and (2) provide software libraries that enable SBML- or PEtab-compatible tools to solve these extended problems, by converting the extended problems to standard SBML and PEtab. We demonstrate these extensions in a real application example of patients in an ICU setting, where they are treated with time-dependent doses of glucose and insulin to achieve healthy blood glucose levels. The optimal control problem is to identify drug doses that keep the patient healthy. Here, one key challenge is that this is an online setting, where new patient data arrives every 15 minutes, and a new drug dose regimen should be proposed on a similar timescale.