COMBINE 2022

The “Computational Modeling in Biology” Network (COMBINE) is an initiative to coordinate the development of the various community standards and formats in systems biology and related fields. COMBINE 2022 will be a workshop-style event with oral presentations, breakout sessions and tutorials. The three meeting days will include talks about the COMBINE standards and associated or related standardization efforts, presentations of tools using these standards, breakout sessions for detailed discussions as well as tutorials.
COMBINE 2022 will be an in-person meeting organised as a satellite to the ICSB 2022 meeting in Berlin.
The daily welcomes, wrap-up and the COMBINE forum will be streamed online. Please register via the following link for the stream: (was hu-berlin.zoom.us/meeting/register/u5EocOGoqzgiE9LPwbHLtC-m_6UTn7LCXx6X)
The COMBINE 2022 meeting is organised by Dagmar Waltemath and Matthias König.
Overview
COMBINE Community meet-up
Two days of breakout sessions and tutorials related to COMBINE standards
Thus 6th & Fri 7th Oct, 8:30am-6pm at Humboldt University of Berlin, Philippstraße 13, House 4
All attendees can suggest breakout sessions for detailed discussions of certain aspects of one or several of the COMBINE standard(s), discussions on metadata and semantic annotations (format-specific or overarching), discussions on the application and implementations of the COMBINE standards, or any other topic relevant for the COMBINE community.
We also solicit applications to host tutorials on tools, standards or resources (or any other topic) which would be relevant for the COMBINE community (see Topics of Interest below). The link to submit suggestions for breakout sessions or tutorials is here.
COMBINE Forum
Single track session with invited and submitted talks
Sat 8th Oct, 8am-1pm at Humboldt University of Berlin, Philippstraße 13, House 4
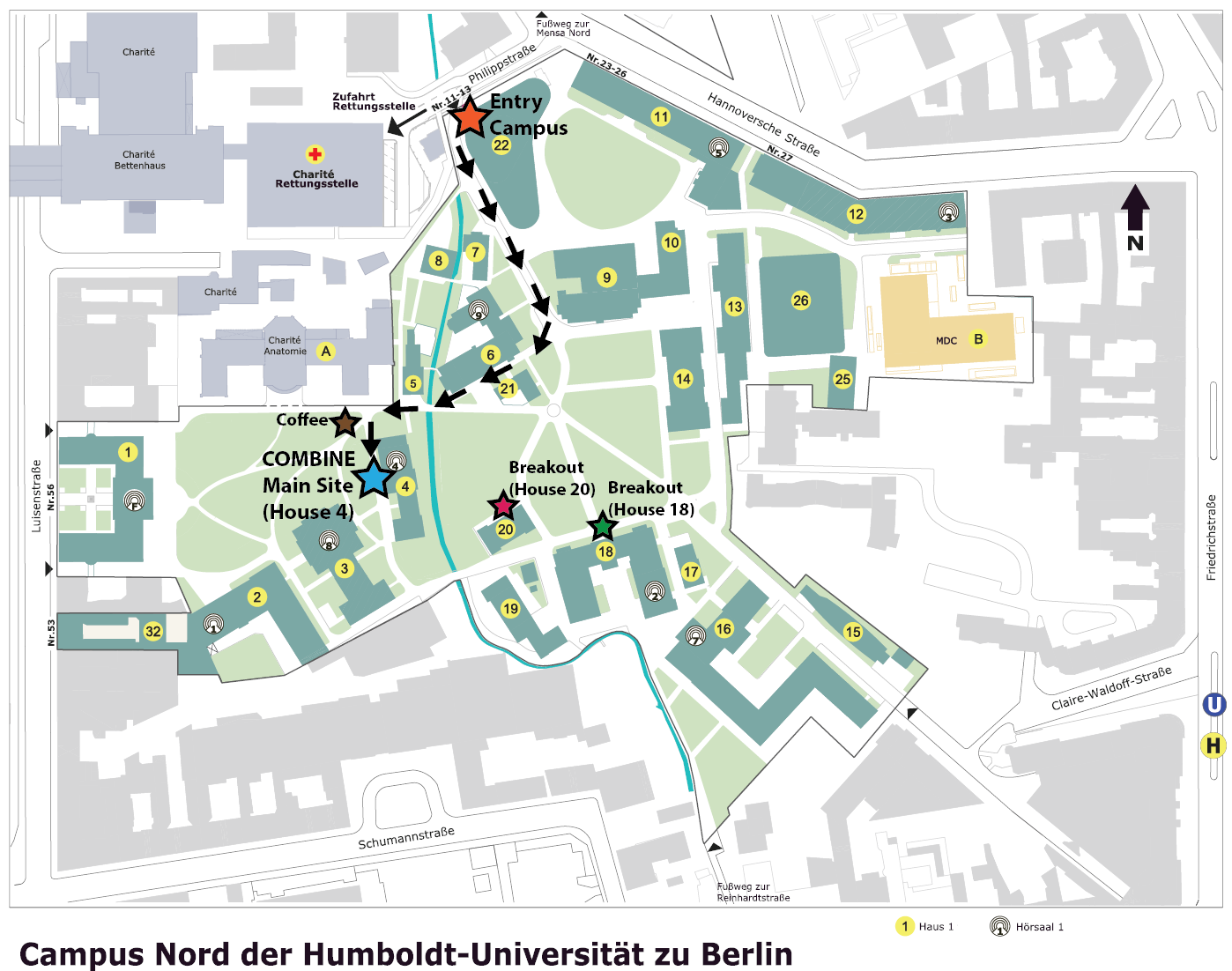
How to get there?
COMBINE2022 is located at Humboldt University of Berlin, Philippstraße 13, House 4
The main entry is at Philippstraße 13. The main event site is in House 4, where the daily welcomes, wrap-ups and the COMBINE Forum will take place. In addition, some breakouts will take place in House 20 and House 18. Please see the following plan for details:
Agenda
Thursday Oct 6 - COMBINE Community meet-up Day 1
| 8:30am | Welcome (Matthias König, House 4, Lecture Hall) |
| 9am | Breakouts & Tutorials 1 Breakouts: - Room 2 (House 4, 012a): SEP 55 (Prashant Vaidyanathan) - Room 1 (House 4, Lecture Hall): SED-ML Level 2: Proposals and Requirements (Lucian Smith) |
| 10:30am | Break / Coffee |
| 11am | Breakouts & Tutorials 2 Tutorial: - Room 3 (House 18, Maut Menten): CovidGraph: Interfaces and Usage (Lea Gütebier) Breakouts: - Room 2 (House 4, 012a): Parametric SVG for SBOL Visual (Thomas Gorochowski) - Room 1 (House 4, Lecture Hall): SED-ML Level 2: Proposals and Requirements |
| 12:30pm | Lunch (self organized) |
| 2pm | Breakouts & Tutorials 3 Tutorial: - Room 3 (House 18, Maud Menten): Clinical trial simulations with the Jinkō modeling platform Breakouts: - Room 2 (House 4, 012a): SBOL 3.1 (Gonzalo Vidal) - Room 1 (House 4, Lecture hall): Curating models and simulations for BioSimulations (David Nickerson) - online: Protocol Activity Modeling Language Standard Working Group (Daniel Bryce) |
| 3:30pm | Break / Coffee |
| 4pm | Breakouts & Tutorials 4 Breakouts: - Room 2 (House 4, 012a): Representation of Site-Variant Libraries (James Diggans) - Room 1 (House 4, Lecture Hall): VSCode-Antimony: A Smart Editor for Developing Models in Systems Biology (Joe Hellerstein/Herbert Sauro) - online: Protocol Activity Modeling Language Standard Working Group |
| 5:30pm | Wrap-up (House 4, Lecture Hall) |
Friday Oct 7 - COMBINE Community meet-up Day 2
| 8:30am | Welcome (Matthias König, House 4, Lecture Hall) |
| 9am | Breakouts & Tutorials 5 Tutorial: - Room 1 (House 4, Lecture Hall): Morpheus: Multi-scale, Multi-cellular, Spatio-temporal Modeling with Hands-on for Your Own Projects Breakouts: - Room 3 (House 20, Seminar Room 1): Managing RDF Packages (Jacob Beal) - Room 2 (House 4, 012a): FROG Analysis - a community standard to foster reproducibility and curation of constraint-based models (Rahuman Sheriff) - Room 4 (House 20, Seminar Room 2): Similarity Computation of Small Molecule Names (Lukrécia Mertová) |
| 10:30am | Break / Coffee |
| 11am | Breakouts & Tutorials 6 Breakouts: - Room 3 (House 20, Seminar Room 1): SBOL Examples (Prashant Vaidyanathan) - Room 2 (House 4, 012a): FAIRness assessment of COMBINE archives (Irina Balaur, Dagmar Waltemath) - Room 1 (House 4, Lecture Hall): MultiCellML – a “to-be” Multi-cellular Modelling language (Jörn Starruß) - Room 4 (House 20, Seminar Room 2): SBML FBC Package Finalization (Matthias König) |
| 12:30pm | Lunch (self organized) |
| 2pm | Breakouts & Tutorials 7 Tutorial: - Room 1 (House 4, Lecture Hall): Creating reproducible biochemical modeling workflows in Python (Part 1) Breakouts: - Room 3 (House 20, Seminar Room 1): RNA Working Group Discussions (Thomas Gorochowski) - Room 2 (House 4, 012a): FAIRness assessment of COMBINE archives - Room 4 (House 20, Seminar Room 2): MultiCellML – a “to-be” Multi-cellular Modelling language (Jörn Starruß) - online: Protocol Activity Modeling Language Standard Working Group (Daniel Bryce) |
| 3:30pm | Break / Coffee |
| 4pm | Breakouts & Tutorials 8 Tutorial: - Room 1 (House 4, Lecture Hall): Creating reproducible biochemical modeling workflows in Python (Part 2) Breakouts: - Room 3 (House 20, Seminar Room 1): RNA Working Group Discussions - Room 4 (House 20, Seminar Room 2): “Data Hazards: framework for thinking ethics in computational systems biology (Susana Roman Garcia)” - Room 2 (House 4, 012a): SBGN general discussion (Adrien Rougny) - online: Protocol Activity Modeling Language Standard Working Group (Daniel Bryce) |
| 5:30pm | Wrap-up (House 4, Lecture Hall) |
| 2:30pm - 5.00pm | - different location: Workshop on statistical inference for dynamical models, PETab (Jan Hasenauer, Daniel Weindl) |
Saturday Oct 8 - COMBINE Forum
| 8am | Poster Session / Coffee |
| 8:45am | Welcome (Dagmar Waltemath and Matthias König) |
| 9am | Invited Talk F.A.I.R. Bayesian Workflows and Model Citation Analysis Nicole Radde, University of Stuttgart |
| 9:30am | Contributed Talks FROG Analysis - a community standard to foster reproducibility and curation of constraint-based models Rahuman Malik Sheriff BioCypher: an ontology-driven framework for flexible harmonisation of large-scale biomedical knowledge graphs Sebastian Lobentanzer Implementing Safe Cross-Document RDF References Jacob Beal The Protocol Activity Modeling Language Jacob Beal |
| 10:30am | Poster Session / Coffee |
| 11am | Invited Talk Documentation of simulation studies — beyond reproducibility Adelinde Uhrmacher, Institute for Visual and Analytic Computing, University Rostock By storing information about a simulation study within a provenance graph the stored information becomes queryable and the dependencies between different modeling and experimenting activities and the used or generated entities become visually accessible. Combined with explicit unambiguous model and simulation experiment specifications such a documentation enhances the reproducibility of the simulation study and the credibility of its products. However, as every documentation of a simulation study, it requires additional effort by the modeler. Therefore, to make the most out of the documentation, how can it be used beyond being a source of information for the modeler about the current or a previous simulation study? In this talk we will discuss the potential, challenges, and experiences in using information stored in a provenance graph to generate and execute simulation experiments during simulation studies automatically. |
| 11:30am | Contributed Talks FAIRDOM: Promoting and Supporting FAIR Data and Model Management in Systems Biology Olga Krebs MeDaX - our vision for bioMedical Data eXploration Judith Wodke Morpheus: Multi-scale, Multi-cellular, Spatio-temporal Modeling Made Easy Jörn Starruß Analyze Parameter Space for Rule-based Models with Simmune AnalyzerFengkai Zhang |
| 12:30pm | Poster Session / Coffee |
| 1pm | Adjourn |
Attendees
86 participants have registered for COMBINE2022 so far.| Name | Affiliation | Attendance |
|---|---|---|
| Mihail Anton | Chalmers University of Technology | Friday 7th October COMBINE Archive and OMEX, FROG, ModeleXchange, Personalized Medicine, SBGN |
| Lara BRUEZIERE | Novadiscovery | Thursday 6th October, Friday 7th October, Saturday 8th October SBGN, SBML, SED-ML |
| Irina Balaur | University of Luxembourg | Thursday 6th October, Friday 7th October, Saturday 8th October COMBINE Archive and OMEX, Personalized Medicine, SBGN |
| Jacob Barhak | Jacob Barhak Analytics | Friday 7th October, Saturday 8th October ModeleXchange, SBML |
| Bryan Bartley | Raytheon BBN | Thursday 6th October, Friday 7th October, Saturday 8th October SBOL, SBOL Visual |
| Florian Bartsch | Institut für theoretische Biologie, Humboldt-Universität zu Berlin | Thursday 6th October, Friday 7th October Personalized Medicine, SBML |
| Jacob Beal | Raytheon BBN | Thursday 6th October, Friday 7th October, Saturday 8th October SBOL, SBOL Visual |
| Frank Bergmann | Heidelberg University, BioQUANT / COS | Thursday 6th October, Friday 7th October, Saturday 8th October COMBINE Archive and OMEX, FROG, PEtab, SBGN, SBML, SED-ML |
| Michael Blinov | UConn Health | Friday 7th October, Saturday 8th October BioPAX, COMBINE Archive and OMEX, ModeleXchange, PEtab, SBGN, SBML, SED-ML |
| Lutz Brusch | Technische Universität Dresden | Friday 7th October, Saturday 8th October COMBINE Archive and OMEX, PEtab, MultiCellML, MorpheusML, SBML-Spatial |
| Daniel Bryce | SIFT, LLC. | Thursday 6th October, Friday 7th October, Saturday 8th October - |
| Lukas Buecherl | University of Colorado Boulder | Thursday 6th October, Friday 7th October, Saturday 8th October CellML, COMBINE Archive and OMEX, Personalized Medicine, SBML, SBOL, SBOL Visual, SED-ML |
| Zsófia Bujtár | Max Delbrück Center for Molecular Medicine in the Helmholtz Association (Berlin) | Friday 7th October, Saturday 8th October SBGN, SBML, DDE-BIFTOOL |
| Famke Bäuerle | University of Tuebingen | Thursday 6th October, Friday 7th October, Saturday 8th October COMBINE Archive and OMEX, FROG, Personalized Medicine, SBGN, SBML, SED-ML |
| Jeremy Cahill | Metamer Labs | Thursday 6th October, Friday 7th October, Saturday 8th October BioPAX, CellML, COMBINE Archive and OMEX, FROG, ModeleXchange, NeuroML, Personalized Medicine, PEtab, SBGN, SBML, SBOL, SBOL Visual, SED-ML |
| Marie Coutelier | Paris Brain Institute | - CellML, COMBINE Archive and OMEX, NeuroML, Personalized Medicine |
| Tobias Czauderna | University of Applied Sciences Mittweida | Friday 7th October, Saturday 8th October SBGN |
| James Diggans | Twist Bioscience | Thursday 6th October SBOL, SBOL Visual |
| Andreas Dräger | Eberhard Karls Universität Tübingen | Thursday 6th October, Friday 7th October, Saturday 8th October COMBINE Archive and OMEX, FROG, SBGN, SBML, SED-ML |
| Dorotea Dudas | HITS gGmbH | Thursday 6th October, Friday 7th October, Saturday 8th October SBML |
| Ibrahim Dulijan | stevens.edu | Thursday 6th October, Friday 7th October, Saturday 8th October SBML, SBOL, SBOL Visual |
| Longxuan Fan | University of Washington | Thursday 6th October SBML |
| Fabian Fröhlich | Harvard Medical School | Friday 7th October, Saturday 8th October BioPAX, PEtab, SBML, SED-ML |
| Alan Garny | University of Auckland | Thursday 6th October, Friday 7th October, Saturday 8th October CellML, COMBINE Archive and OMEX, SED-ML |
| Robert T. Giessmann | Institute for globally distributed open research and education (IGDORE) | - BioPAX, COMBINE Archive and OMEX, SBML, SBOL, SBOL Visual, SED-ML, thermodynamic data, openTECR |
| Mose Giordano | UCL | Thursday 6th October, Friday 7th October, Saturday 8th October SBML |
| Thomas Gorochowski | University of Bristol | Thursday 6th October, Friday 7th October, Saturday 8th October SBOL, SBOL Visual |
| Alexander Gower | Chalmers University of Technology | Thursday 6th October, Friday 7th October, Saturday 8th October BioPAX, CellML, COMBINE Archive and OMEX, FROG, ModeleXchange, PEtab, SBGN, SBML, SED-ML |
| Niklas Gröne | Universität Konstanz | Thursday 6th October, Friday 7th October, Saturday 8th October - |
| Roy Gusinow | University of Bonn | Thursday 6th October, Friday 7th October, Saturday 8th October NeuroML, PEtab, SBML |
| Lea Gütebier | Department of Medical Informatics, Institute for Community Medicine, University Medicine Greifswald | Thursday 6th October, Friday 7th October, Saturday 8th October CellML, Personalized Medicine, SBML, SED-ML |
| Jan Hasenauer | University of Bonn | Thursday 6th October, Friday 7th October, Saturday 8th October ModeleXchange, Personalized Medicine, PEtab, SBGN, SBML, SED-ML |
| Joseph Hellerstein | eScience Institute, University of Washington | Thursday 6th October SBML, SED-ML |
| Joseph Hellerstein | University of Washington | Thursday 6th October PEtab, SBML, SED-ML |
| Ron Henkel | University Medicine Greifswald | Thursday 6th October, Friday 7th October, Saturday 8th October CellML, COMBINE Archive and OMEX, ModeleXchange, SBGN, SBML, SBOL, SED-ML |
| Henning Hermjakob | EMBL-EBI | Friday 7th October, Saturday 8th October BioPAX, COMBINE Archive and OMEX, FROG, ModeleXchange, SBML |
| Sebastian Höpfl | Institute for Systems Theory and Automatic Control (IST) | Thursday 6th October, Friday 7th October, Saturday 8th October COMBINE Archive and OMEX, Personalized Medicine, PEtab, SBML |
| Johann Fredrik Jadebeck | IBG-1 Forschungszentrum Jülich | Thursday 6th October, Saturday 8th October FROG, SBML |
| João Júnior | Centro de Tecnologia Canavieira | Thursday 6th October, Friday 7th October, Saturday 8th October CellML, ModeleXchange, Personalized Medicine, SED-ML |
| Miroslav Kratochvíl | Luxembourg Centre for Systems Biomedicine | Saturday 8th October FROG, SBML |
| Olga Krebs | Heidelberg Institute for Theoretical Studies | Thursday 6th October, Friday 7th October, Saturday 8th October Personalized Medicine, PEtab, SBML, SBOL, SBOL Visual, SED-ML |
| Adrian Köller | Institut für theoretische Biologie, Humboldt-Universität zu Berlin | Thursday 6th October, Friday 7th October SBML |
| Matthias König | Humboldt-University Berlin | Thursday 6th October, Friday 7th October, Saturday 8th October COMBINE Archive and OMEX, FROG, Personalized Medicine, PEtab, SBML, SED-ML |
| Paul Lang | University of Oxford; Julia Computing | Saturday 8th October Personalized Medicine, PEtab, SBML |
| Eva Liu | University of Washington Bioengineering Sys-Bio Lab | Thursday 6th October - |
| Sebastian Lobentanzer | University Hospital Heidelberg | Thursday 6th October, Friday 7th October, Saturday 8th October BioPAX, Personalized Medicine, PEtab, SBGN, SBML |
| Augustin Luna | Harvard Medical School | Thursday 6th October, Friday 7th October, Saturday 8th October BioPAX, SBGN |
| Karin Lundengård | Auckland Bioengineering Institute | Thursday 6th October, Friday 7th October - |
| Steve Ma | NVIDIA | Thursday 6th October SBML |
| Simon Merkt | University of Bonn | Thursday 6th October, Friday 7th October, Saturday 8th October NeuroML, PEtab, SBGN, SBML |
| Lukrécia Mertová | Heidelberg Institute for Theoretical Studies HITS gGmbH | Thursday 6th October, Friday 7th October, Saturday 8th October BioPAX, ModeleXchange, Personalized Medicine, SBML, SBOL |
| Goksel Misirli | Keele University | Thursday 6th October, Friday 7th October SBOL, SBOL Visual |
| Ion Moraru | UConn Health | Thursday 6th October, Friday 7th October COMBINE Archive and OMEX, SBML, SED-ML |
| Sébastien Moretti | SIB Swiss Institute of Bioinformatics | Thursday 6th October, Friday 7th October SBML |
| Chris Myers | University of Colorado Boulder | Thursday 6th October, Friday 7th October, Saturday 8th October SBOL, SBOL Visual, SBML, SED-ML |
| Tung Nguyen | EMBL-EBI | Thursday 6th October, Friday 7th October, Saturday 8th October BioPAX, CellML, COMBINE Archive and OMEX, FROG, ModeleXchange, Personalized Medicine, PEtab, SBGN, SBML, SED-ML |
| David Nickerson | Auckland Bioengineering Institute, University of Auckland | Thursday 6th October, Friday 7th October, Saturday 8th October CellML, COMBINE Archive and OMEX, ModeleXchange, Personalized Medicine, PEtab, SED-ML |
| Dilan Pathirana | University of Bonn | Thursday 6th October, Friday 7th October, Saturday 8th October PEtab, SBML |
| Clemens Peiter | IRU MLS, LIMES, University of Bonn | Thursday 6th October, Friday 7th October PEtab, SBGN, SBML |
| Sathish Periyasamy | Queensland Brain Institute | Friday 7th October, Saturday 8th October CellML, NeuroML, Personalized Medicine, SBGN, SBML |
| Fronth Nyus Pi | Quantum4Life Sensonomiqs | Thursday 6th October, Friday 7th October, Saturday 8th October CellML, COMBINE Archive and OMEX, NeuroML, SBML, SBOL |
| Veronica Porubsky | University of Washington | Thursday 6th October, Friday 7th October, Saturday 8th October NeuroML |
| Nicole Radde | University of Stuttgart | Friday 7th October, Saturday 8th October COMBINE Archive and OMEX, ModeleXchange, Personalized Medicine, PEtab, SBML |
| Karthik Raman | Indian Institute of Technology Madras | Thursday 6th October, Friday 7th October, Saturday 8th October FROG |
| Gianmarco Rasi | - | Thursday 6th October, Friday 7th October, Saturday 8th October SBML |
| Susana Roman Garcia | University of Edinburgh | Thursday 6th October, Friday 7th October, Saturday 8th October BioPAX, CellML, COMBINE Archive and OMEX, ModeleXchange, NeuroML, Personalized Medicine, PEtab, SBGN, SBML, SBOL, SBOL Visual, SED-ML |
| Adrien Rougny | Independent researcher (former AIST) | Friday 7th October, Saturday 8th October SBGN |
| Herbert Sauro | University of Washington | Thursday 6th October, Friday 7th October, Saturday 8th October COMBINE Archive and OMEX, SBML, SED-ML |
| Falk Schreiber | University of Konstanz | Thursday 6th October, Friday 7th October, Saturday 8th October SBGN, SBML, SBOL Visual |
| James Scott-Brown | University of Edinburgh | Thursday 6th October, Friday 7th October SBOL, SBOL Visual |
| Vinoo Selvarajah | iGEM Foundation | Thursday 6th October, Friday 7th October SBOL, SBOL Visual |
| Woosub Shin | Auckland Bioengineering Institute, University of Washington | - SBML |
| Lucian Smith | University of Washington | Thursday 6th October, Friday 7th October, Saturday 8th October CellML, COMBINE Archive and OMEX, FROG, PEtab, SBML, SED-ML |
| Jörn Starruß | TU Dresden | Thursday 6th October, Friday 7th October, Saturday 8th October PEtab, SBML, MultiCellML |
| Melanie Stefan | Medical School Berlin | Thursday 6th October, Friday 7th October, Saturday 8th October COMBINE Archive and OMEX, Personalized Medicine, SBGN, SBML |
| Abel Szkalisity | University of Helsinki | Thursday 6th October, Friday 7th October, Saturday 8th October BioPAX, SBML |
| Baishakhi Tikader | Indian Institute of Bombay, India | Friday 7th October SBML |
| Krishna Kumar Tiwari | EMBL-EBI | Thursday 6th October, Friday 7th October, Saturday 8th October FROG, ModeleXchange, Personalized Medicine, SBGN |
| Matej Troják | Masaryk university | Thursday 6th October, Friday 7th October, Saturday 8th October SBML |
| Adelinde Uhrmacher | University of Rostock | Saturday 8th October CellML, COMBINE Archive and OMEX, SBGN, SBML, SED-ML |
| Prashant Vaidyanathan | Oxford Biomedica | Thursday 6th October, Friday 7th October SBOL |
| Paola Vera-Licona | UConn Health | Friday 7th October BioPAX, Personalized Medicine, SBML |
| Gonzalo Vidal | ICOS, Newcastle University | Thursday 6th October, Friday 7th October, Saturday 8th October SBOL, SBOL Visual |
| Dagmar Waltemath | University Medicine Greifswald | Thursday 6th October, Friday 7th October, Saturday 8th October COMBINE Archive and OMEX, ModeleXchange, Personalized Medicine, SBML, SED-ML, FAIR model indicators |
| Daniel Weindl | Helmholtz Munich | Thursday 6th October, Friday 7th October, Saturday 8th October PEtab, SBML, SED-ML |
| St. Elmo Wilken | Heinrich Heine University | Saturday 8th October - |
| Pia Wilsdorf | University of Rostock | Saturday 8th October COMBINE Archive and OMEX, PEtab, SED-ML |
| Fengkai Zhang | NIAID/NIH | Thursday 6th October, Friday 7th October, Saturday 8th October COMBINE Archive and OMEX, ModeleXchange, SBGN, SBML, SED-ML |
| kristina gruden | NIB | Saturday 8th October CellML, SBGN, SBML, SBOL |